GeneticData
09_Tree_of_life.RmdA quick wrap-up of the git stuff from last time
We’ll install a package called usethis, which contains
some helpful programming tools for the weeks ahead:
install.packages("usethis")We’ll use this to create a personal access token for github. This ensures we don’t have to retype our password every time we need to access GitHub.
usethis::create_github_token() And we will use this token to access our account via RStudio.
credentials::set_github_pat("PAT here")Now, refresh your R session.
Once this is done, we can check that this worked like so:
usethis::git_sitrep()The Tree of Life
First, we need to do some installs.
install.packages("devtools")
devtools::install_github("ropensci/rotl")
#if (!requireNamespace("BiocManager"))
# install.packages("BiocManager")
#BiocManager::install()
# Install package dependencies
#BiocManager::install("Biostrings")
#BiocManager::install("biomaRt")
#install.packages("biomartr", dependencies = TRUE)Now, load your packages:
Open Tree
First, we’re going to look for some data. Discuss with your neighbor a clade you like and want to build a tree of. For example, perhaps I really like hominids.
library(rotl)
apes <- c("Pongo", "Pan", "Gorilla", "Hoolock", "Homo")
(resolved_names <- rotl::tnrs_match_names(apes))## search_string unique_name approximate_match score ott_id is_synonym
## 1 pongo Pongo FALSE 1 417949 FALSE
## 2 pan Pan FALSE 1 417957 FALSE
## 3 gorilla Gorilla FALSE 1 417969 FALSE
## 4 hoolock Hoolock FALSE 1 712902 FALSE
## 5 homo Homo FALSE 1 770309 FALSE
## flags number_matches
## 1 2
## 2 sibling_higher 2
## 3 sibling_higher 1
## 4 1
## 5 extinct_inherited, sibling_higher 1Put the names of five genera into the function. If you want, you can use mine below, or pick new ones you prefer.
ants <- c("Martialis", "Atta", "Ectatomma", "Tatuidris", "Aneuretus", "Xymmer", "Aenictus")
(resolved_names <- rotl::tnrs_match_names(ants))## search_string unique_name approximate_match score ott_id is_synonym flags
## 1 martialis Martialis FALSE 1 334634 FALSE
## 2 atta Atta FALSE 1 709964 FALSE
## 3 ectatomma Ectatomma FALSE 1 162845 FALSE
## 4 tatuidris Tatuidris FALSE 1 150177 FALSE
## 5 aneuretus Aneuretus FALSE 1 425343 FALSE
## 6 xymmer Xymmer FALSE 1 4583488 FALSE
## 7 aenictus Aenictus FALSE 1 169919 FALSE
## number_matches
## 1 1
## 2 1
## 3 1
## 4 1
## 5 1
## 6 1
## 7 1Try to view them on a tree:
tr <- tol_induced_subtree(ott_ids = ott_id(resolved_names))## Progress [----------------------------------] 0/43 ( 0%) ?sProgress [================================] 43/43 (100%) 0s ## Warning in collapse_singles(tr, show_progress): Dropping singleton nodes with
## labels: mrcaott1682ott35311, mrcaott1682ott34821, mrcaott34821ott950983,
## Aneuretinae ott393261, mrcaott4706ott5525, mrcaott5525ott8038,
## mrcaott8038ott8685, mrcaott8685ott36581, mrcaott8685ott56535,
## mrcaott8685ott72729, mrcaott8685ott86888, mrcaott86888ott129302, Attini
## ott344333, mrcaott114767ott114779, mrcaott114779ott339738,
## mrcaott114779ott116619, mrcaott116619ott148996, mrcaott148996ott1071522,
## mrcaott148996ott150184, mrcaott148996ott339763, mrcaott148996ott409299,
## mrcaott48019ott688185, Ectatomminae ott327970, mrcaott48019ott162057,
## mrcaott7438ott5553505, mrcaott7438ott7439, mrcaott7439ott165217,
## mrcaott7439ott15430, mrcaott15430ott120483, mrcaott15430ott150187,
## mrcaott15430ott169913, mrcaott17752ott98880, mrcaott17752ott150176,
## mrcaott150176ott1037289, mrcaott150176ott4539262, Agroecomyrmecinae ott504610,
## mrcaott98886ott152924, mrcaott152924ott452719, mrcaott152924ott739132,
## mrcaott152924ott503313, mrcaott503313ott928530, mrcaott928530ott6280946,
## Martialinae ott334635
plot(tr)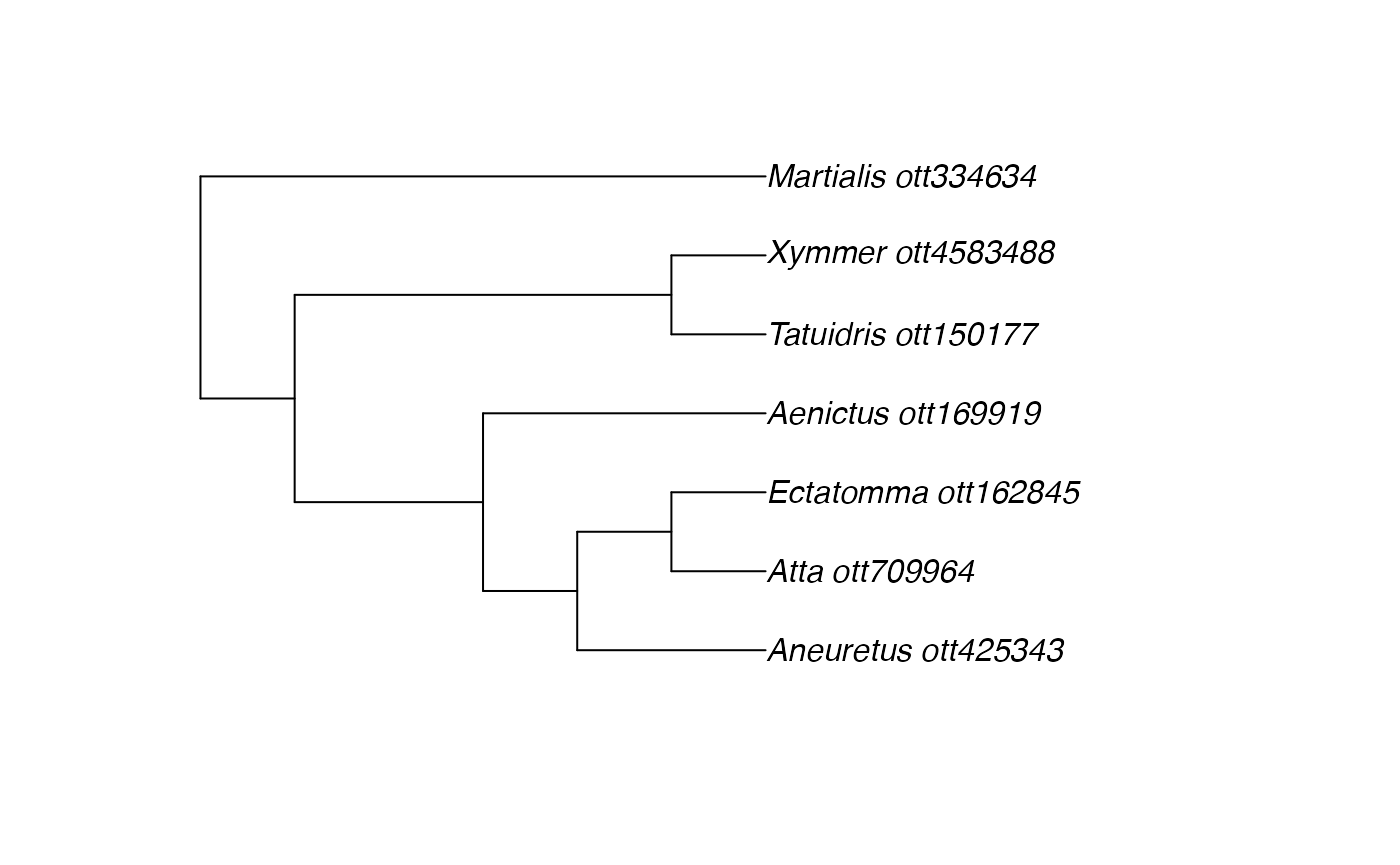
I’m a little surprised by this tree.
Looking for data
We’re going to try to build a tree of these taxa from GenBank Data. Much like accessing NOAA data, we will need to make an API Key. Go here and register.
Now that we have that key, we can access NCBI:
library(rentrez)
entrez_search(db = "Nucleotide", term="Ectatomma", api_key ="96b569c049fe3d3055c5b747112dfec84308")## Entrez search result with 707 hits (object contains 20 IDs and no web_history object)
## Search term (as translated): "Ectatomma"[Organism] OR Ectatomma[All Fields]Holy moley that’s a lot of data. If we do that for every taxon, we’ll end up with a mess. Why don’t we try to filter it a bit.
Ectatomma <- entrez_search(db = "Nucleotide", term="Ectatomma AND 2018:2019[PDAT]", retmax = 100, api_key ="96b569c049fe3d3055c5b747112dfec84308")So there is a good amount of data for our Ectatomma ants. We’re going
to download some. And this gets a little tricky. The first thing we will
do is try the entrez_fetch command:
entrez_fetch(db="nuccore", id = Ectatomma$ids,
rettype="fasta", retmax=1)## [1] ">MK759606.1 Ectatomma ruidum voucher YB-BCI64481 cytochrome oxidase subunit 1 (COI) gene, partial cds; mitochondrial\nATATTTCATCTTTGCTATCTGAACTGGTTTAATTGGTTCATCTATAAGTATAATTATTCGATTAGGGCTA\nGGAACTTGCGGTTCTATTATTAATAATGACCAAGTTTTTAATTCTATTGTTACAGCTCATGCATTTATTA\nTAATTTTTTTCATGGTTATACCATTTATAATTGGAGGATTTGGAAATTTTTTAGTGCCTTTAATATTAGG\nATCTCCTGATATAGCCTACCCACGAATAAATAATATAAGATTTTGACTCCTTCCTCCCTCTATTATACTT\nTTAACTTTAAGAAGATTAATTAGTAGAGGAGTGGGAACAGGATGAACAGTTTATCCTCCCCTCGCATCTA\nGTGTTTTCCATAGAGGTTCGTCTGTTGATCTAGCTATTTTTTCATTACATATTGCAGGAATATCATCAAT\nTTTAGGGGCTACTAATTTTATTTCAACTATTATAAATATACACCATAAAAATATAACTATAGATAAAATT\nCCTCTTTTAGTTTGAGCTATTATAATTATTGCAATTTTACTACTTTTATCGTTACCTGTATTAGCTGGTG\nCTATTACAATACTTTTAACTGACCGAAATTTAAATACTTTATTCTTTGATCCTTCAGGAAGTGGGGATCC\nTATTCTTTATCAACATTTATTT\n\n"Look at the output - does it look like we have output for every ID?
Let’s try the below. If you’ve forgotten, this is called a
for loop. It means that for each entry in the ids vector,
we will download a sequence from GenBank.
for( id in Ectatomma$ids) {
recs <- entrez_fetch(db="nuccore", id = id,
rettype="fasta", retmax=1)
cat(recs, file="Ect.seqs.fasta", append=TRUE)
}Now, let’s look at this file. What do we notice about it? What information is in this file?
Before we move on, we’re going to try one other thing. Try modifying one of the two chunks above (HINT: you will have to rerun both the search and the fetch) in order to only get data from the mitochondrial COI gene.
All right, let’s move on to trying to write a function that will return all IDs for all of the ants.
search_year <- function(ant){
query <- paste0(ant, " AND ", "2016:2019[PDAT]")
search_returns <- c(entrez_search(db="Nucleotide", term=query, retmax=20,api_key ="96b569c049fe3d3055c5b747112dfec84308"))
return(search_returns)
}
ants <- as.vector(resolved_names$unique_name)
labels <- sapply(ants,search_year, USE.NAMES = T)Have a look at ?sapply. Talk to a partner and see if you can figure out what sapply does.
So what is the problem with this output? It’s hard to parse - let’s make it easier to pull data for one taxon at a time.
search_year <- function(ant){
search_returns = c()
query <- paste0(ant," AND ", "2019:2022[PDAT]")
search_returns[ant] <- c(entrez_search(db="Nucleotide", term=query, retmax=20,api_key ="96b569c049fe3d3055c5b747112dfec84308"))
return(search_returns)
}
ants <- as.vector(resolved_names$unique_name)
labels <- sapply(ants,search_year, USE.NAMES = T)There’s one remaining problem, though. Have a look at your outputs. How do you think we can fix this one? Let’s have a look at when papers related to Xymmer were published:
entrez_fetch(db="pmc", term="Xymmer", id= "1103332937", retmax=20, rettype= "text", api_key ="96b569c049fe3d3055c5b747112dfec84308")Run this as-is and take a look at the outputs.
search_year <- function(ant){
search_returns = c()
query <- paste0(ant, "[Organism]", " AND 18s")
search_returns[ant] <- c(entrez_search(db="Nucleotide", term=query, retmax=1,api_key ="96b569c049fe3d3055c5b747112dfec84308"))
return(search_returns)
}
ants <- as.vector(resolved_names$unique_name)
labels <- sapply(ants,search_year, USE.NAMES = F)Now, we’re going to get the genetic data.
search_seq <- function(labels){
recs <- entrez_fetch(db="nuccore", id = labels,
rettype="fasta", retmax=1)
cat(recs, file="Ants.seqs.fasta", append=TRUE)
return(recs)
}
seq_returns <- sapply(labels, search_seq, USE.NAMES = T)Excercise:
18s is an easy one. Let’s try and pull another common gene for phylogenetics, COX1.
Make a new cell and modify our search_year code to pull COX1 data. What are the most immediate problems you notice?
Try resolving the issue. Can you pinpoint which part of our query gives us trouble?
Just for fun
Take a quick look at this output - it’s a little neat. We’re not going to go far into building phylogenetic trees - that’s a topic for Systematics. Here’s a quick look at these data.
install.packages("ape")##
## The downloaded binary packages are in
## /var/folders/54/9kd8nf1x4fnft0ymvb11qmc80000gn/T//Rtmp9thPyS/downloaded_packages
s18 <- ape::read.dna("Ants.seqs.fasta", format="fasta")These data are referred to as “unaligned” - this means we haven’t
assigned homology at the molecular level. The process of doing this is
called sequence alignment. Let’s take a look at doing that. We’ll
install a package called msa, for Multiple Sequence
Alignment.
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("msa")## 'getOption("repos")' replaces Bioconductor standard repositories, see
## 'help("repositories", package = "BiocManager")' for details.
## Replacement repositories:
## CRAN: https://cran.rstudio.com/## Bioconductor version 3.19 (BiocManager 1.30.25), R 4.4.1 (2024-06-14)## Warning: package(s) not installed when version(s) same as or greater than current; use
## `force = TRUE` to re-install: 'msa'## Loading required package: Biostrings## Loading required package: BiocGenerics##
## Attaching package: 'BiocGenerics'## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs## The following objects are masked from 'package:base':
##
## anyDuplicated, aperm, append, as.data.frame, basename, cbind,
## colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
## get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
## match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
## Position, rank, rbind, Reduce, rownames, sapply, setdiff, table,
## tapply, union, unique, unsplit, which.max, which.min## Loading required package: S4Vectors## Loading required package: stats4##
## Attaching package: 'S4Vectors'## The following object is masked from 'package:utils':
##
## findMatches## The following objects are masked from 'package:base':
##
## expand.grid, I, unname## Loading required package: IRanges## Loading required package: XVector## Loading required package: GenomeInfoDb##
## Attaching package: 'Biostrings'## The following object is masked from 'package:base':
##
## strsplitThe process of assessing homology at the molecular level typically involves a model of sequence evolution.
library(Biostrings)
mySequence <- readDNAStringSet("../Ants.seqs.fasta")## Warning in .Call2("fasta_index", filexp_list, nrec, skip, seek.first.rec, :
## reading FASTA file ../Ants.seqs.fasta: ignored 2293 invalid one-letter sequence
## codes
myAlignment <- msa(mySequence, "ClustalOmega")## using Gonnet
output_seq <- msaConvert(x = myAlignment, type = "phangorn::phyDat")And we can even estimate a little phylogenetic tree:
## Loading required package: ape##
## Attaching package: 'ape'## The following object is masked from 'package:Biostrings':
##
## complement##
## Phylogenetic tree with 7 tips and 5 internal nodes.
##
## Tip labels:
## KU672072.1 Martialis heureka voucher CAS:ENT:0106181 abdominal A (abd-A) gene, partial cds, KU672072.1 Martialis heureka voucher CAS:ENT:0106181 abdominal A (abd-A) gene, partial cds, KU672072.1 Martialis heureka voucher CAS:ENT:0106181 abdominal A (abd-A) gene, partial cds, KU672072.1 Martialis heureka voucher CAS:ENT:0106181 abdominal A (abd-A) gene, partial cds, KU672072.1 Martialis heureka voucher CAS:ENT:0106181 abdominal A (abd-A) gene, partial cds, KU672072.1 Martialis heureka voucher CAS:ENT:0106181 abdominal A (abd-A) gene, partial cds, ...
##
## Unrooted; includes branch lengths.Or using parsimony: