Functions and repeatability
April Wright; Borrows heavily from Ethan White’s semester long biology course
05_Functions.RmdMultiple data sets: By Column
So far, we have looked at a situation in which we have one spreadsheet. But often times, you may want to work across multiple spreadsheets. For example, perhaps your lab has historically collected one spreadsheet per experiment or data collection. Or you’re using Excel workbook style. Let’s look at examples of each of these.
First, let’s take a look at a situation in which we have our data split across two sheets. This commonly happens if we have different devices or software recording data. In this case, for the butterflies dataset, we have one data sheet for the temperatures and one for the cumultative preceptitation. This is because those data points were collected with two different data logging devices that each record separately.
Let’s take a peek at these data, and see if we can combine them to get a more complete picture of our data.
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.5
## ✔ forcats 1.0.0 ✔ stringr 1.5.1
## ✔ ggplot2 3.5.1 ✔ tibble 3.2.1
## ✔ lubridate 1.9.3 ✔ tidyr 1.3.1
## ✔ purrr 1.0.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
temp <- read_csv("../data/Butterfly_data.csv")## Rows: 144 Columns: 5
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (1): ButterflySpecies
## dbl (4): Year, Day, SpringTemp, SummerTemp
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
rainfall <- read_csv("../data/Butterfly_moisture.csv")## Rows: 144 Columns: 4
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (1): ButterflySpecies
## dbl (3): Year, Day, Cumulative_Rain
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Take a quick look at these data. In what situation might it be useful to record days in this manner?
We have a common column in this dataset, which makes joining them together easy. Sort of.
butterflies_joined <- full_join(rainfall, temp, by= c("ButterflySpecies", "Year", "Day"), multiple = "all", relationship = "many-to-many")If you take a look at these data, we now have all the columns present in the new data. This is a little unsatisfying. You might notice that some Species-Year-Day combinations are the same. If you were collecting data of this sort, you might consider assigning each observation a unique ID in the way that the surveys dataset does.
But still, we can now do some neat things with these data.
ggplot(butterflies_joined, aes(x = Day, y = SpringTemp, color = Cumulative_Rain)) + geom_point()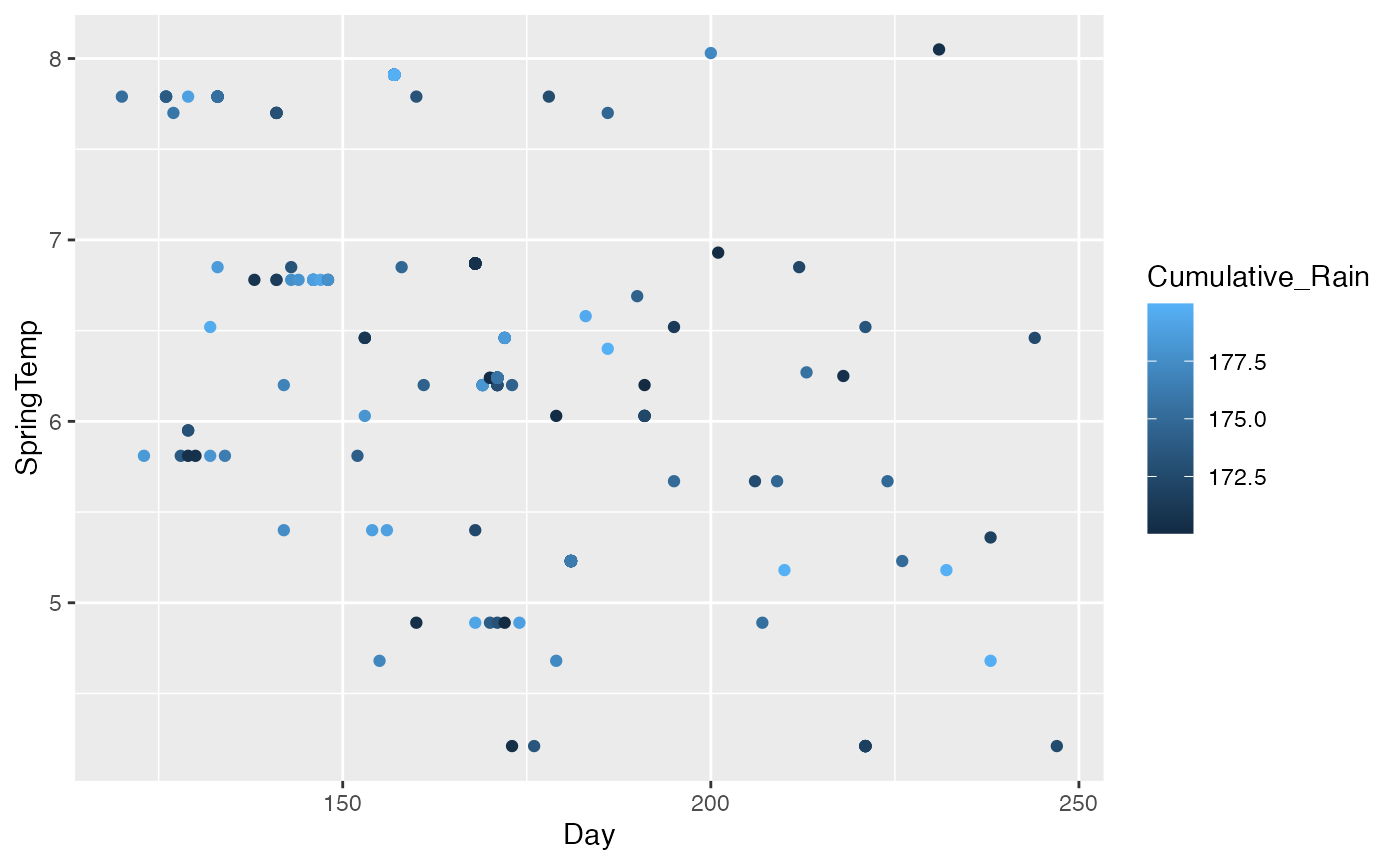
Multiple Datasets: By Row
In the previous example, we had a dataset where each observation was split across two datasheets. However, sometimes we might have experiments that are split across multiple datasheets. Perhaps, for example, you record data in one sheet for the control and another for the treatment. This is pretty common in different kinds of experiments - you wouldn’t want someone to accidentally record treatment data with the control group.
In this case, we’re going to take a look at some of your classmate’s phylogenetic data. It’s exported in a format that is common across phylogenetics and in other software packages. It is called ‘tab-separated’, as opposed to comma-separated. We import it a little differently.
log1 <- read_delim("../data/Kaitie_log1.log", delim = "\t")## Rows: 601 Columns: 224
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: "\t"
## dbl (224): Iteration, Posterior, Likelihood, Prior, alpha_morpho, branch_rat...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
log2 <- read_delim("../data/Kaitie_log2.log", delim = "\t")## Rows: 601 Columns: 224
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: "\t"
## dbl (224): Iteration, Posterior, Likelihood, Prior, alpha_morpho, branch_rat...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.What I want you to do is take a minute and scroll the columns - does anything jump out as a way to distinguish one log from another? Are any columns different?
In this case, we may need to add a column to the dataset specifying what the dataset is.
log1 <- log1 %>%
mutate(Treatment = "Control")
log2 <- log2 %>%
mutate(Treatment = "Treatment")
combined <- rbind(log1, log2)If you look at this object, it now has our datasets stacked on top of one another. This might not seem intutitively better, but it enables more complex grouping and plotting. For example, perhaps we want per-group means displayed in an easy-to-see table.
transform_table <- combined %>%
select(Treatment, starts_with("t[")) %>%
group_by(Treatment) %>%
summarize()
transform_table## # A tibble: 2 × 1
## Treatment
## <chr>
## 1 Control
## 2 TreatmentOr we can plot:
##
## Attaching package: 'reshape2'## The following object is masked from 'package:tidyr':
##
## smiths
data_mod <- combined %>%
select(Treatment,starts_with("t[") ) %>%
melt(id.vars='Treatment')
ggplot(data_mod) +
geom_boxplot(aes(x=variable, y=value, color=Treatment)) 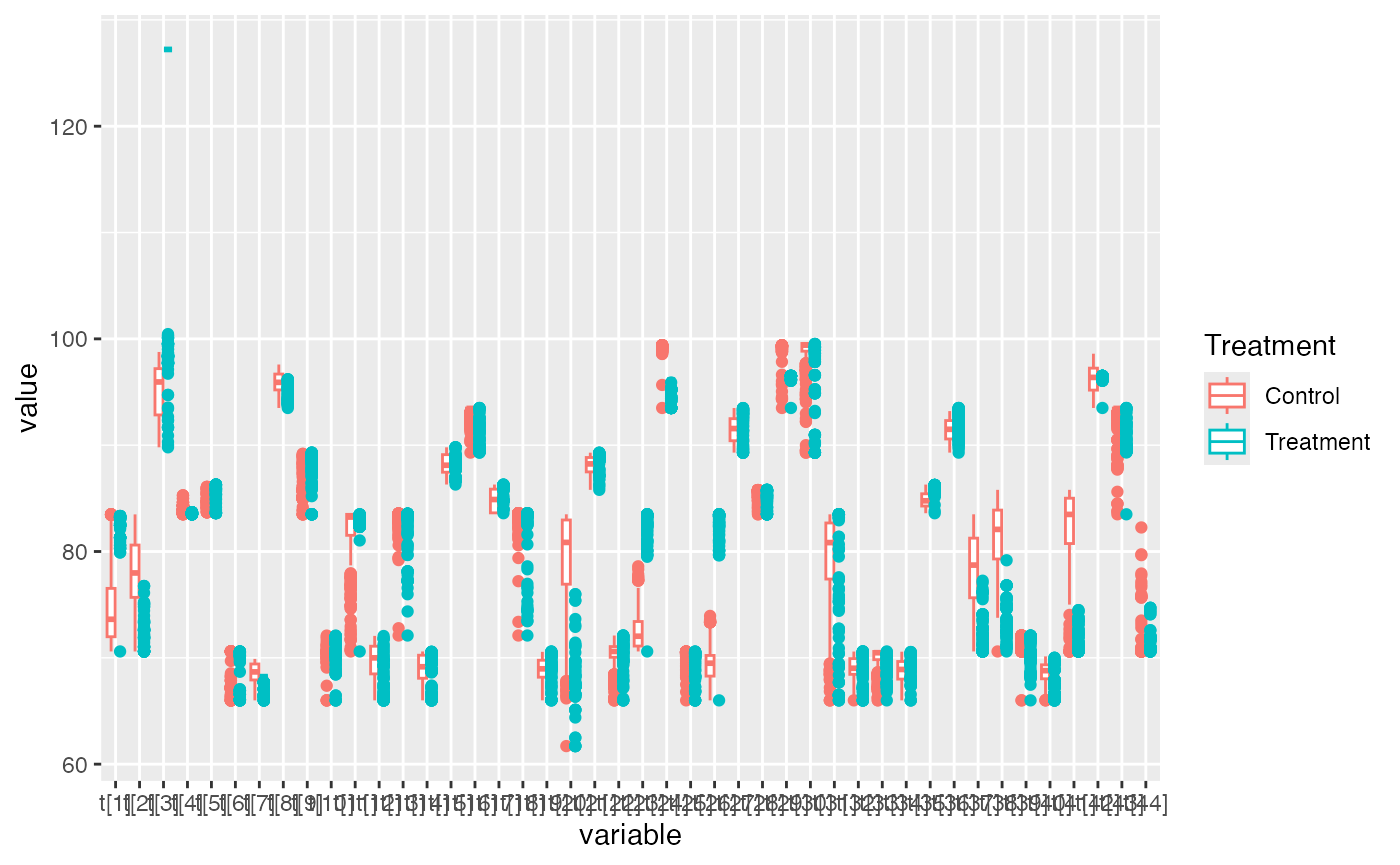 ## Multiple Data Sheets: Workbook edition
## Multiple Data Sheets: Workbook edition
A common spreadsheet organization is the Workbook. Let’s look at an example. YOu can see how this is appealing, and it tends to be fairly common in both field and experimental work. We’re going to download a couple packages for working with this sort of data:
install.packages("googlesheets4")##
## The downloaded binary packages are in
## /var/folders/54/9kd8nf1x4fnft0ymvb11qmc80000gn/T//RtmpRu0tRO/downloaded_packages
install.packages("readxl")##
## The downloaded binary packages are in
## /var/folders/54/9kd8nf1x4fnft0ymvb11qmc80000gn/T//RtmpRu0tRO/downloaded_packagesGoogleSheets4 is a library for working directly with data from Google. It can be very handy if you’re working collaboratively with others on projects.
The gs4_deauth function allows you to access public
datasheets without entering your password. We’ll use it to download the
data straight into R:
raw_data <- read_sheet("https://docs.google.com/spreadsheets/d/1uSPKCKFpNMmbbKvqf0W4P2SPSB3xu68LOJkdFOEaFNM/edit?usp=sharing")## ✔ Reading from Butterly_data_by_Site.## ✔ Range Utrecht.Oh no! Who sees a problem? We need to get sheets individually. So the convenience of Google Drive is somewhat counterbalanced by the rudimentary nature of the tools.
utrecht <- read_sheet("https://docs.google.com/spreadsheets/d/1uSPKCKFpNMmbbKvqf0W4P2SPSB3xu68LOJkdFOEaFNM/edit?usp=sharing", sheet = c("Utrecht"))## ✔ Reading from Butterly_data_by_Site.## ✔ Range ''Utrecht''.
erlangen <- read_sheet("https://docs.google.com/spreadsheets/d/1uSPKCKFpNMmbbKvqf0W4P2SPSB3xu68LOJkdFOEaFNM/edit?usp=sharing", sheet = c("Erlangen"))## ✔ Reading from Butterly_data_by_Site.## ✔ Range ''Erlangen''.
zurich <- read_sheet("https://docs.google.com/spreadsheets/d/1uSPKCKFpNMmbbKvqf0W4P2SPSB3xu68LOJkdFOEaFNM/edit?usp=sharing", sheet = c("Zurich"))## ✔ Reading from Butterly_data_by_Site.## ✔ Range ''Zurich''.
combined <- rbind(utrecht, erlangen, zurich)Another way to tackle this is readxl This is a library
that generates some more sophisticated outputs from Excel workbooks. For
example, this same workbook can be read in this way:
workbook <- readxl::read_excel("../data/Butterly_data_by_Site.xlsx", sheet = "Zurich")But the thing to know is that if data are in separate worksheets, R will assume them to be separate, and you must have a protocol for merging them, if you desire to do so.
Functions enable repeatability
- How many of you have done something so far in the semester, and then
forgotten it later? Let’s discuss how to make code more permenant,
reusable and readable
- Informative variable names
- Consistent indentation and line spacing
- Good commenting
- Functions
- Write reusable code.
- Concise and modular script
- Functions with general structure
Understandable chunks
- Human brain can only hold ~7 things in memory at a given time.
- Write programs that don’t require remembering more than ~7 things at once.
- What do you know about how
sum(1:5)works internally?- Nothing.
- What do you think it does?
- Test your idea. Were you right?
- All functions should work as a single conceptual chunk, labeled in a logical way so that you don’t have to remember what it does.
Reuse
- Want to do the same thing repeatedly?
- Inefficient & error prone to copy code
- Even worse to rewrite it every time!
- If it occurs in more than one place, it will eventually be wrong somewhere.
- Functions are written to be reusable.
Function basics
function_name <- function(inputs) {
output_value <- do_something(inputs)
return(output_value)
}calc_shrub_vol <- function(length, width, height) {
volume <- length * width * height
return(volume)
}- Creating a function doesn’t run it.
- Call the function with some arguments.
calc_shrub_vol(0.8, 1.6, 2.0)
shrub_vol <- calc_shrub_vol(0.8, 1.6, 2.0)- Walk through function execution (using debugger)
- Call function
- Assign 0.8 to length, 1.6 to width, and 2.0 to height inside function
- Calculate volume
- Send the volume back as output
- Store it in
shrub_vol
- Treat functions like a black box.
- Can’t access a variable that was created in a function
> widthError: object 'width' not found
- ‘Global’ variables can influence function, but should not.
- Very confusing and error prone to use a variable that isn’t passed in as an argument
- Can’t access a variable that was created in a function
Exercise:
Type the name of a function that we have already used in class to view the source code of the function. Can you tell what the expected inputs and outputs of the function are?
Default arguments
- Defaults can be set for common inputs.
calc_shrub_vol <- function(length = 1, width = 1, height = 1) {
volume <- length * width * height
return(volume)
}
calc_shrub_vol()
calc_shrub_vol(width = 2)
calc_shrub_vol(0.8, 1.6, 2.0)
calc_shrub_vol(height = 2.0, length = 0.8, width = 1.6)Named vs unnamed arguments
- When to use or not use argument names
calc_shrub_vol(height = 2.0, length = 0.8, width = 1.6)Or
calc_shrub_vol(2.0, 0.8, 1.6)- You can always use names
- Value gets assigned to variable of that name
- Remember the 7 things we can hold in memory?
- If not using names then order determines naming
- First value is
height, second value islength… - If order is hard to remember use names
- First value is
- In many cases there are a lot of optional arguments
- Convention to always name optional argument
Combining Functions
Each function should be single conceptual chunk of code
Functions can be combined to do larger tasks in two ways
Calling multiple functions in a row
est_shrub_mass <- function(volume){
mass <- 2.65 * volume^0.9
}
shrub_mass <- est_shrub_mass(calc_shrub_vol(0.8, 1.6, 2.0))
library(dplyr)
shrub_mass <- calc_shrub_vol(0.8, 1.6, 2.0) %>%
est_shrub_mass()- Calling functions from inside other functions
- Allows organizing function calls into logical groups
est_shrub_mass_dim <- function(length, width, height){
volume = calc_shrub_vol(length, width, height)
mass <- est_shrub_mass(volume)
return(mass)
}
est_shrub_mass_dim(0.8, 1.6, 2.0)Consistency
We ran into some trouble Tuesday. I had defined each of my functions a couple times. And in so doing, I ended up with extra arguments to some functions and none to others.
calc_vol <- function(len = 1, width = 10, height = 1){
volume <- len * width * height
return(volume)
}
calc_mass <- function(vol){
mass <- 2.65 * vol^.9
return(mass)
}
calc_den <- function(len, width, height){
vol <- calc_vol(len, width, height)
mass <- calc_mass(len)
density <- mass/vol
return(density)
}
calc_den(len = 2, width = 3, height = 4 )## [1] 0.2060448More complex functions
So far, we’ve looked at situations where we are providing one or a few arguments to a function. But what about situations where we want to be able to use functions to process our data in more complex ways?
We may want to select data from a data frame inside a function. We can do that like so:
surveys <- read_csv("../data/surveys.csv")## Rows: 35549 Columns: 9
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (2): species_id, sex
## dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
remove_nas <- function(data){
clean <- data %>%
na.omit(data) %>%
select(weight)
return(clean)
}
remove_nas(surveys)## # A tibble: 30,676 × 1
## weight
## <dbl>
## 1 40
## 2 48
## 3 29
## 4 46
## 5 36
## 6 52
## 7 8
## 8 22
## 9 35
## 10 7
## # ℹ 30,666 more rowsBut is this a good thing? This function will allow us to select one and only one column. We call this practice hardcoding. It makes our code inflexible - what if we want other data? We would have to edit the code itself. In general, we want to make as few changes to our code as possible. Every change introduces the chance to make a mistake.
So, we can change our code a little bit to make it more flexible. We can, for example, make it so that the user tells the function what code they want. We do this by placing the column in the function body in curly braces. This tells R to prepare to accept user input. See the below:
remove_nas <- function(data, column){
clean <- data %>%
na.omit(data) %>%
select({{column}})
return(clean)
}
remove_nas(surveys, hindfoot_length)## # A tibble: 30,676 × 1
## hindfoot_length
## <dbl>
## 1 35
## 2 37
## 3 34
## 4 35
## 5 35
## 6 32
## 7 15
## 8 21
## 9 36
## 10 12
## # ℹ 30,666 more rowsAfter you have run the above, you will see that we can now choose a column based on what we provide in the function call. Very handy!
Exercise:
We may want multiple columns of data. How can we make a function flexible to multiple columns? There are two possible solutions to this issue. Look at each of the below and see if you can figure out how they work. Discuss with a partner the pros and cons of each solution.
#Solution One
remove_nas <- function(data, column, column1){
clean <- data %>%
na.omit(data) %>%
select({{column}}, {{column1}})
return(clean)
}
remove_nas(surveys, hindfoot_length, weight)## # A tibble: 30,676 × 2
## hindfoot_length weight
## <dbl> <dbl>
## 1 35 40
## 2 37 48
## 3 34 29
## 4 35 46
## 5 35 36
## 6 32 52
## 7 15 8
## 8 21 22
## 9 36 35
## 10 12 7
## # ℹ 30,666 more rowsTesting
- How can we know what we’ve done is good?
- How could you verify that a number is right?
- What about our density? What must be true of it?
- Generic vs. specific
- What about non-numerics?
- Write some function to clean data.
Functions from class
calc_density <- function(length, width, height){
volume <- calc_vol(length, width, height)
mass <- calc_mass(volume)
density <- mass/volume
if (density < volume){
return(density)
} else {
print("Density impossibly large! Check your math.")
}
}
calc_density <- function(length, width, height){
volume <- calc_vol(length, width, height)
mass <- calc_mass(volume)
density <- mass/volume
density <- round(density, 3)
if (is.double(density)){
return(density)
} else {
print("Density impossibly large! Check your math.")
}
}
data_cleaning <- function(filepath){
data_raw <- read_csv(filepath)
data_clean <- data_raw %>%
drop_na()
if (sum(is.na(d_c)) == 0){
return(data_clean)
} else {
print("NAs still present!")
}
}